Overview
Colorectal cancer is the third most aggressive cancer worldwide. Polyps (the main biomarker of the disease), are characterized through colonoscopy procedures. Nonetheless, due to high appearance variability, up to 25\% of polyps are missed, impacting the survival rate (8% of survival probability at the fourth stage). Additionally, real clinical scenarios have to address the poor patient preparation and intestinal folds that may be confused with polyp masses, where polyp observations appear in less than 1% of the frames.
COLON Dataset 2023 aims to discuss new guidelines and perspectives of computational approaches to operate in real clinical scenarios where colonoscopy sequences include: water bubbles, food waste; polyps at different stages, size, morphology (sessile or pedunculated), and the corresponding biopsy results. The researchers are invited to propose computational strategies that characterize polyps by their segmentation and localization taking advantage of real clinical recordings that include full intestinal background and some sequences.
About
The COLON Dataset
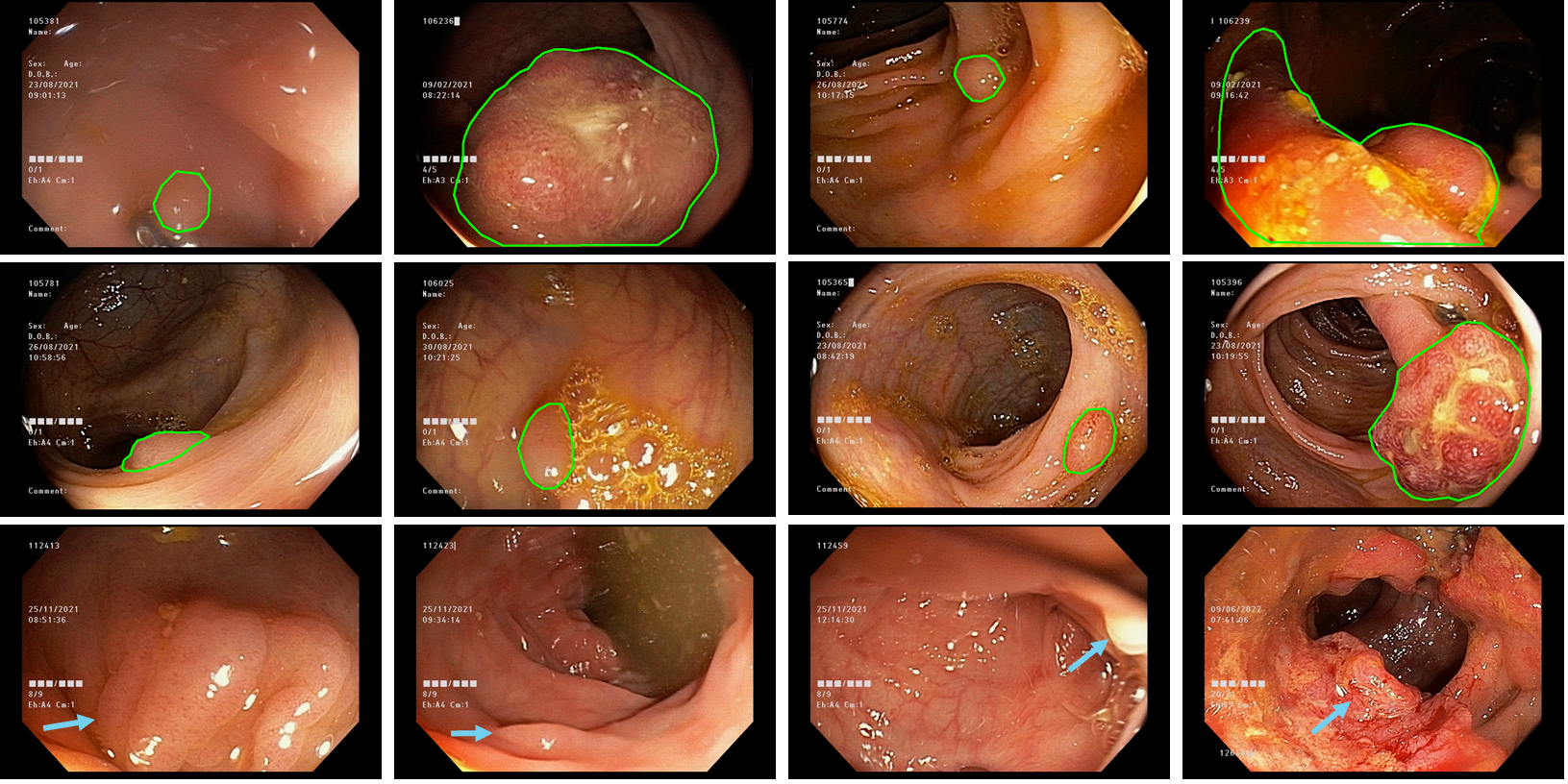
The COLON Dataset collects the major number of polyp and non-polyp frame observations. This dataset recovered samples from 30 colonoscopy video sequences with polyps, from typical procedures with a huge amount of intestinal background frames together with polyp findings with high visual and shape variability. These findings were labeled by an expert gastroenterologist with more than 13 years of experience. For training, the colonoscopy sequences were labeled every 10 frames, while for the test dense labeling (every frame of the sequence) was carried out at each frame of the sequence. The dataset aims to compare computational strategies around the polyp localization and segmentation tasks and discuss new guidelines and perspectives of computational approaches to operate in real clinical scenarios against the medical expert criteria.
How to use the dataset?
If you are interested in participating, you are invited to download the training set in the data section, which includes 20 and 5 colonoscopy videos with and without polyp (more than 1800 and 60000 frames, respectively).
The website will perform the evaluation of the proposed methods via docker methodology. To complete a successful participation, participants are encouraged to submit an abstract, describing the employed method. All results will be publicly available in results section, only the top 10 results will be displayed on the website.
Each participant team are encouraged to submit their method as a poster or journal in any other conference after ISBI 2023 ends. Eventually, submissions will be potentially compiled for a high-impact journal paper to summarise and present the findings.
Motivation
Colorectal cancer (CRC) is the third most common cancer and the second most deadly cancer worldwide. Polyps (the main CRC biomarker) detection and shape characterization are key factors in the patient’s survival rate. Polyps are typically observed from colonoscopy procedures, but their detection and respective characterization require exhaustive observations, taking an average of 20 to 30 minutes. Additionally, related procedure challenges (high polyp appearance variability due to constant illumination changes, artifacts in the intestinal tract, and abrupt camera movements) difficult the proper polyp characterization reporting a miss detection rate between 6-25% during the clinical procedure. These critical issues affect the early diagnosis impacting directly the survival rate, i.e., patients at the fourth stage have an 8% of survival probability.
Data
The proposed COLON Dataset was elaborated in collaboration with Instituto de Gastroenterología y Hepatología del Oriente - IGHO S.A.S. and the BIVL2ab (Biomedical, Imaging and Vision Learning Laboratory) from Universidad Industrial de Santander in Bucaramanga, Colombia. This dataset includes colonoscopies recorded with Olympus Evis Excera III 190 colonoscopy, with a spatial resolution of 480×720 and a temporal resolution of 30 fps. The dataset contains 30 long sequences with polyps and 10 long sequences without polyps and at least three sequences have two polyps. The polyp annotations were carried out by an expert gastroenterologist with more than 13 years of experience.
Participate
The data distribution, registration and automatic evaluation will be handled by BIVL2ab team. The following link let you register into the challenge and await for the administrators confirmation in participating.
It is highly recommended to use your institutional email address for the registration. Please note that while you can use non-institutional emails (e.g. Gmail, Hotmail, Yahoo, etc.) your request might be refused depending on the license model of the dataset and its intended use. The registration form will ask you for previous experience and interest in participating on the challenge.
After registration
You will receive an email with an acceptance or decline in your team participating telling you the reasons. We cordially ask you for your patience while waiting for a response from BIVL2ab team. Later on other mail, you will find an unique link to download the dataset with a 7 day expire date. In case you didn't get to download the data, please send us an email to our email to generate a new link.
Submission
How to submit?
Click the following link to enter in submission website. The credentials to login are send to the team leader email at the time of acceptance.
After you login with your credentials, you will be able to upload your model contained in a zip file following the next steps:
- Select the "COLON 2023" option in the "Challenge to submit" selector.
- Select the zip file that contains your Dockerfile and model weights (Remember to follow the docker template).
- Press the "Submit" button and wait for your docker to be correctly uploaded (The status is displayed on the right window).
- For a visual explanation, you can see the following video.
Docker template guidelines
In order to make a successful submission you can download and follow the comments instructions in the docker template zip which contains a Dockerfile and evaluate.py files with their respective instructions. The following are general rules to submit your solution in the challenges:
- Your zip must contain the Dockerfile and evaluate.py which have defined elements that mustn't be modified in order to run.
- You can attach inside the zip your model weights and model structure in any ".py", ".h5", ".pth", etc. There are no rules for the files contained in the zip file, except for Dockerfile and evaluate.py files.
- The image used in Dockerfile must be available in Docker Hub Repository.
- In case of any error during the execution of the uploaded zip, it will be shown in the status window on the right.
- You can upload as many solutions you want but only the best will be stored in the server. All the others solutions will have their results only.
Organizers

Franklin Sierra
Dataset Leader
Franklin Sierra is currently a PhD student at Universidad Industrial de Santander in the Computer Engineering and Informatics School. He is part of the Biomedical Imaging, Vision, and Learning Laboratory research group. His main interests are related to computer vision and bioinformatics applications.

Jair Ruiz
Expert Gastroenterologist
Jair Ruiz is a Specialist in Gastroenterology and Digestive Endoscopy and graduated from the Pontificia Universidad Javeriana. He is also an Internal Medicine Specialist from the Universidad Industrial de Santander. Currently, he works as a gastroenterologist, manager, and legal representative of the IGHO S.A.S.

Lina Ruiz
Research assistant
Lina Ruiz is an MSc researcher in the Computer Engineering and Informatics School. She is part of the Biomedical Imaging, Vision, and Learning Laboratory research group. Her main interests are related to medical image processing and deep learning.

Fabio Martínez
Director and Advisor
Fabio Martínez is currently a full-time professor at Universidad Industrial de Santander on the Computer Engineering and Informatics School. He is the director of BIVL2ab research group (Biomedical Imaging, Vision and Learning Laboratory). His principal interests are related to video processing, machine learning, computer vision, and medical imaging processing.



