News
Overview
Stroke represents the second leading cause of mortality worldwide. The key component for immediate diagnosis is the localization (over CT scans) and delineation of lesions ( over MRI studies). The lesions are nonetheless poorly delineated, only visible at advanced stages, and analysis uses manual delineation.
APIS Challenge 2023 pretends to be a workshop that introduces a paired dataset of CT and ADC studies. The researchers are invited to propose computational strategies that approach paired data, during training, and deal with lesion segmentation over CT onset sequences. During training will be available annotated paired sequences (from one expert), and resultant segmentation for testing will be compared regarding two experts.
Dates
| Registration opening | |
| Release and distribution of paired CT-ADC cases | |
| Release and distribution of annotations by radiologist | |
| Workshop to prepare for docker submission | |
| Submission opening and release of docker templates for submission | |
| Deadline for submissions | |
| Presentation and releasing of results for all teams at workshop challenge day | |
| Enabling the challenge website with automated registration and participants creation. | |
| Coming soon... | Presentation of methods and results in challenge website |
About
The APIS Challenge
The APIS Challenge is a workshop dedicated to stroke segmentation from paired (CT-ADC) dataset. For training, 60 paired studies with CT and ADC image modalities are released, with an associated delineation from an expert radiologist. Regarding the testing stage, a total of 40 studies will be used as validation.
We encourage participants to learn projections from ADC and CT sequences and to carry out predicted delineations over CT studies. The challenge claims to capture the generalization capability of proposed approaches, by measuring resultant segmentations from two independent expert radiologists with more than five years of experience. Optionally, authors may generate synthetic ADC images and submit them to the challenge to receive quantitative results from reconstruction metrics.
How to participate in APIS challenge?
If you are interested in participating, you are invited to download the training set in data section, which includes CT and ADC modalities of 60 stroke patients.
The website will perform the evaluation of the proposed methods via docker methodology. To complete a successful participation, participants are encouraged to submit an abstract, describing the employed method. All results will be publicly available in results section, only the top 10 results will be displayed in the website.
Each participant team are encouraged to submit their method as a poster or journal in any other conference after ISBI 2023 ends. Eventually, submissions will be included in the workshops post-proceedings and potentially compiled for a high-impact journal paper to summarise and present the findings.
Prizes
- The three top-performing teams are eligible to participate in a joint publication with the committee sumitting to an Open Access Journal. There is a fixed maximum of two authors per team. The committee will also invite teams that submit particularly novel solutions to join as co-authors.
- The 10 top-performing teams will have the opportunity to present their results at ISBI 2023
Motivation
Strokes are the second leading cause of mortality worldwide, being responsible for the major adult disability in developed countries. A dramatic projection estimates that one in four people over 25 years will suffer a stroke. Ischemic stroke, related to blood vessel occlusion, is the most prevalent condition (80% of all cases). Even worse, this stroke has an associated high morbidity risk. The immediate localization, delineation and quantification of the lesion are fundamental to determining the diagnosis and consequent clinical intervention.
Typically, the first sequence taken is a CT without contrast (NCCT), which mainly allows triage between strokes and other pathologies. However, such sequences have poor contrast, low sensitivity and specificity with respect to ischemia, and hypo-attenuation of lesions is time-dependent. Hence, clinical protocols include diffusion weighted MRI sequences, such as ADC (apparent diffusion coefficient) to properly characterize and quantify stroke lesions, allowing among others, to display ischemic penumbra properly and ischemic core. Such characterization is however tedious, time-consuming (around 15 minutes in each case) and prone to errors, introducing an inherent radiologist bias.
This challenge introduces the first stroke segmentation challenge (APIS) with an associated paired (CT-ADC) publicly available dataset. The main approach is to capture the generalization capability of proposed approaches, by measuring resultant segmentations against two independent expert radiologists with more than five years of experience. The proposals should receive CT inputs and generate lesion segmentations.
Data
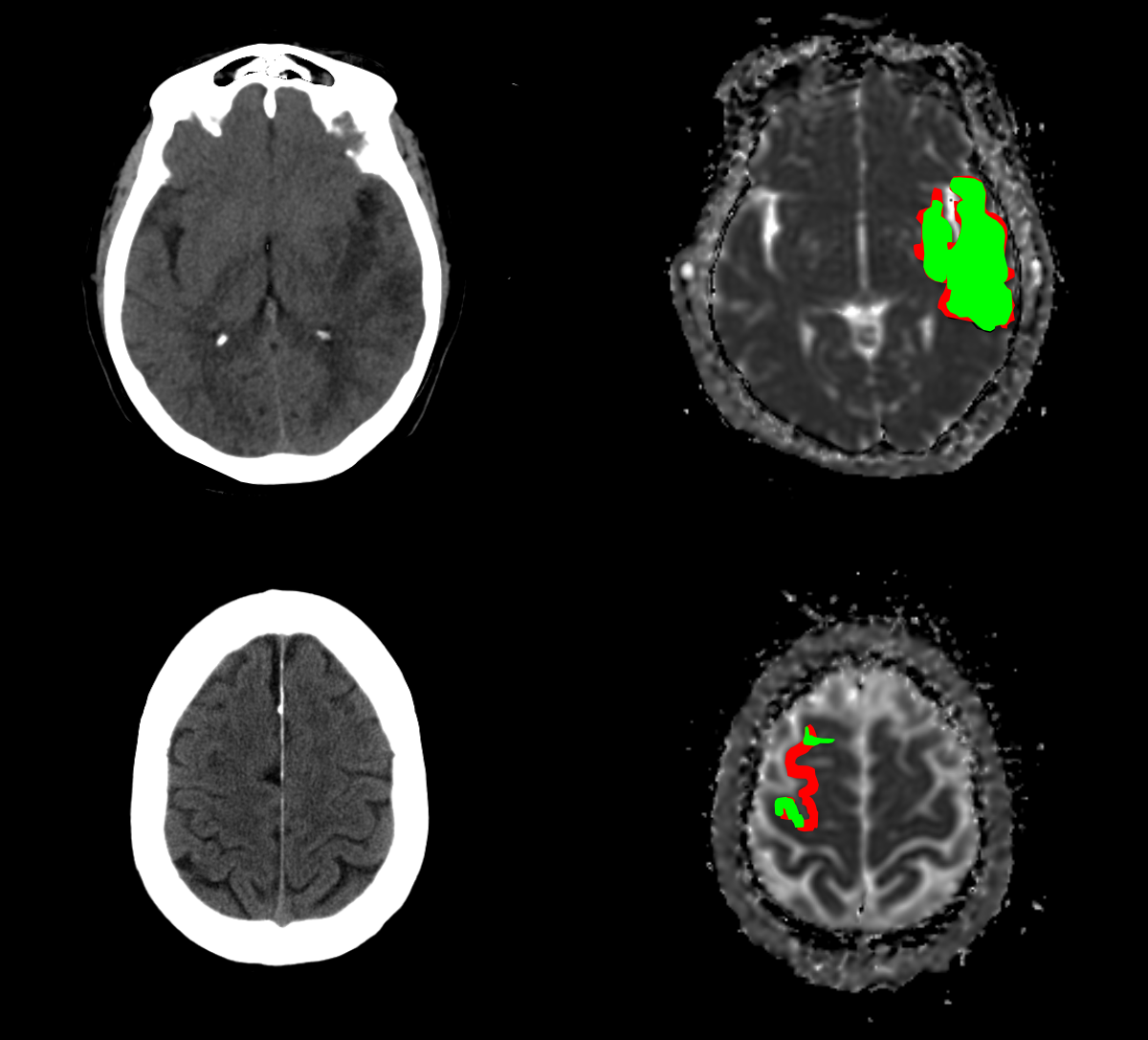
The proposed challenge was elaborated by BIVL2ab (Biomedical, Imaging and Vision Learning Laboratory) from Universidad Industrial de Santander, in collaboration with FOSCAL (Fundacion Oftalmologica de Santander) and FOSUNAB (FOSCAL and Universidad Autonoma de Bucaramanga) clinics. The studies were captured using: two CT scanners, namely i) a Toshiba Aquilion 64 TSX-101 and ii) a Toshiba Aquilion one TSX-301/ac scanner, with 64 and 320 detectors, respectively. Also, one Toshiba Vantage Titan MRI scanner was employed for acquisition of different MRI modalities. Such scanners are located at FOSCAL and FOSUNAB clinics in Bucaramanga, Colombia. Spatial resolution for NCCT images was 512x512 px, and 256x256 for most of diffusion sequences.
The annotation of ischemic lesions was carried out by two neurointerventional radiologists with more than five years of experience. The annotation protocol involved the examination of DWI B-1000 and ADC images to delineate the damaged tissue in the brain. To achieve a more accurate diagnosis, experts had access to clinical variables previous to the manual delineation of each case. The lesion annotation process was performed on the ADC images by each radiologist individually using the MRICroGL software. Furthermore, the associated NCCT studies were skull-stripped and co-registered w.r.t. labeled ADC sequences. The following figure shows a preview of the image modalities and the respective segmentation mask. The delineated lesions show high variability in shape, size and number of consecutive slices where the lesion occurs.
Download APIS Dataset
The data distribution is restricted only for accepted user team. In the following link your registered email address is required to obtain the download link. If you are not registered please go to participate section and register your team. It is highly recommended to use your institutional email address for the registration.
This dataset contains non-contrast computed tomographies (NCCT) and apparent diffusion coefficient (ADC) images from patients diagnosed with ischemic stroke. By doing so, you agree to the following terms of use:
- Users of this data must abide by the Data Usage Policy and the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/) under which it has been published. Attribution should include the reference to the following paper:
"Gómez, Santiago, et al. "APIS: a paired CT-MRI dataset for ischemic stroke segmentation-methods and challenges." Scientific Reports 14.1 (2024): 20543."
-
DATA USAGE POLICY
You agree to reference the recommended bibliographic citation(s) in any publication that employs these resources. These references may be updated till 2024, and you agree to receive emails indicating those updates. This dataset requires the data consumer to comply with access regulations imposed both by law and by the data repository, and to conform to codes of conduct that are generally accepted in higher education and scientific research for the exchange of knowledge and information.
-
PRIVACY OF RESEARCH SUBJECTS
The Dataset is considered anonymized for every subject as established in Resolution Nº 008430 of October 4, 1993, which establishes the scientific, technical and administrative norms for health research in Colombia. The results of the processing of such information may be disclosed for scientific purposes, through presentations at conferences or publications in scientific journals, with the required protection of subject identities. However, by linking this dataset to other, not de-identified datasets it may be theoretically possible to identify single RESEARCH SUBJECTS of this dataset. To more information, please complete the next form, download the APIS Dataset Informed Consent and submit it.
Participate
The data distribution, registration and automatic evaluation will be handled by BIVL2ab team. The following link let you register into the challenge and await for the system confirmation.
It is highly recommended to use your institutional email address for the registration. Please note that while you can use non-institutional emails (e.g. Gmail, Hotmail, Yahoo, etc.) your request might be refused depending on the license model of the dataset and its intended use. The registration form will ask you for previous experience and interest in participating on the challenge.
After registration
You will receive an email with an acceptance or decline in your team participating telling you the reasons. We cordially ask you for your patience while waiting for a response from BIVL2ab team. Later on other mail, you will find an unique link to download the dataset with a 7 day expire date. In case you didn't get to download the data, please send us an email to our email to generate a new link.
Submission
How to submit?
Click the following link to enter in submission website. The credentials to login are send to the team leader email at the time of acceptance.
After you login with your credentials, you will be able to upload your model contained in a zip file following the next steps:
- Select the "APIS ISBI 2023" option in the "Challenge to submit" selector.
- Select the zip file that contains your Dockerfile and model weights (Remember to follow the docker template).
- Press the "Submit" button and wait for your docker to be correctly uploaded (The status is displayed on the right window).
- For a visual explanation, you can see the following video.
Docker template guidelines
In order to make a successful submission you can download and follow the comments instructions in the docker template zip which contains a Dockerfile and evaluate.py files with their respective instructions. The following are general rules to submit your solution in the challenges:
- Your zip must contain the Dockerfile and evaluate.py which have defined elements that mustn't be modified in order to run.
- You can attach inside the zip your model weights and model structure in any ".py", ".h5", ".pth", etc. There are no rules for the files contained in the zip file, except for Dockerfile and evaluate.py files.
- The image used in Dockerfile must be available in Docker Hub Repository.
- In case of any error during the execution of the uploaded zip, it will be shown in the status window on the right.
- You can upload as many solutions you want but only the best will be stored in the server. All the others solutions will have their results only.
Organizers

Santiago Gómez
Challenge Leader
Santiago Gómez is a computer science graduate student at Universidad Industrial de Santander. He is part of BIVL2ab research group (Biomedical Imaging, Vision and Learning Laboratory). His major interests are in deep learning and medical image applications.

Daniel Eduardo Mantilla García
Expert Radiologist
Daniel Eduardo Mantilla García is a Diagnostic and Interventional Neuroradiologist and the director of the Interventional Neuroradiology department at FOSCAL Clinic. Currently coursing his fifth year as PhD candidate at the mathematics school at Montpellier University. His research interests include topics about Interventional Neuroradiology.

Gustavo Garzón
Research assistant
Gustavo Garzón is currently a PhD candidate and adjunct professor at Universidad Industrial de Santander (Colombia). He is part of BIVL2ab research group (Biomedical Imaging, Vision and Learning Laboratory). His major interests are computer vision, deep learning and medical imaging processing.

Edgar Rangel Pieschacon
Laboratory Chief and Challenge Support
Edgar Rangel Pieschacon is a computer science doctoral student at Universidad Industrial de Santander. He is the laboratory chief of BIVL2ab research group (Biomedical Imaging, Vision and Learning Laboratory). His major interests are machine learning, applied statistics and applications related to medical imaging processing.

Andres Felipe Ortiz
Expert Radiologist
Andres Felipe Ortiz is a Interventional Neuroradiologist fellow at the Universidad Autónoma of Bucaramanga (UNAB) - Clinica Ardila Lulle and a diagnostic radiologist who graduated from the Universidad Nacional de Colombia. His research interests include topics in neurointerventionism.

Daniela D Vera
Research assistant
Daniela D Vera is a Physician who graduated from the Universidad Autónoma de Bucaramanga (UNAB). Currently belongs to the Radiology and Diagnostic Imaging group of FOSCAL clinic. His research interests include interventional radiology topics and radiology and diagnostic images.

Fabio Martínez
Director and Advisor
Fabio Martínez is currently a full-time professor at Universidad Industrial de Santander on the Computer Engineering and Informatics School. He is the director of BIVL2ab research group (Biomedical Imaging, Vision and Learning Laboratory). His principal interests are related to machine learning, computer vision, and medical imaging processing.

Paul Anthony Camacho López
Clinical Director
Paul Anthony Camacho López is the director of reasearch and development department at FOSCAL Clinical Center (Fundacion Oftalmologica de Santander). He is a surgeon, epidemiologist, health care administrator and researcher. He obtained his master's degree in epidemiology in the Universidad Industrial de Santander and graduated as a physician from the Universidad Nacional de Bogotá.






